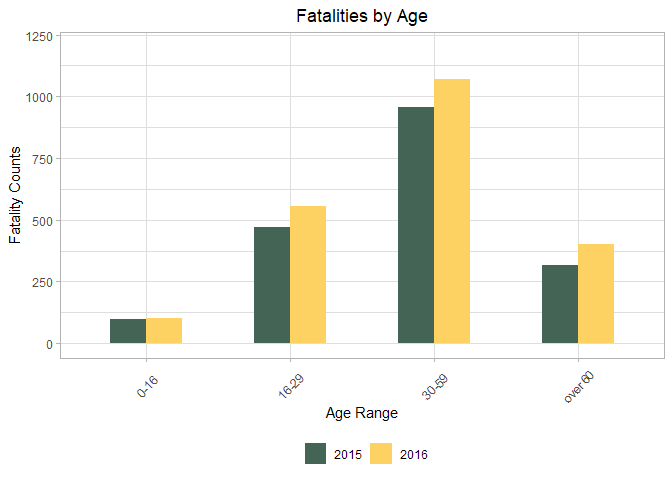This is a note for the Fatality Analysis Reporting System (FARS) data. A hit-and-run data wrangling was conducted during the internship at AAA Foundation for Traffic Safety on summer, 2018.
Outline
FARS data
- functions in tidyverse
- dplyr::filter
- dplyr::select
- dplyr::distinct
- dplyr::left_join
- dplyr::gather and dplyr::spread
- magrittr::%>% (pipe operator)
- base functions
- table
- merge
- paste0
- assign
- cut
- sapply
- ggplot2
- geom_bar
1. Fatality Analysis Reporting System (FARS) data set
FARS is a nationwide census data system regarding fatal injuries
suffered in motor vehicle traffic crashes across the United States,
including 50 States, District of Columbia, and Pureto Rico.
FARS
website
Where you can download raw data from:
FARS Raw Data –
ftp://ftp.nhtsa.dot.gov/FARS
As of the date of the internship, it has records from 1975 to 2017.
Manuals and documents of FARS:
FARS Manuals
What are included in FARS?
143 data elements (variables) that characterize three types of data:
- Crash data
- Vehicles data
- Person data
Read the data file into R
Following examples using only the data from 2015 to 2016.
## Load necessary library
library(tidyverse)
## Read CSV file, using function read.csv("path", header, sep)
for(X in 2015:2016){
p0 <- paste0("./FARS/person_", X, ".csv")
p1 <- paste0("per_", X, sep = "")
v0 <- paste0("./FARS/vehicle_", X, ".csv")
v1 <- paste0("veh_", X, sep = "")
a0 <- paste0("./FARS/accident_", X, ".csv")
a1 <- paste0("acc_", X, sep = "")
assign(p1, read.csv(p0, header = TRUE, sep = ","))
assign(v1, read.csv(v0, header = TRUE, sep = ","))
assign(a1, read.csv(a0, header = TRUE, sep = ","))
}
Take a look at the data
head(as_data_frame(per_2015))
## # A tibble: 6 x 68
## STATE ST_CASE VE_FORMS VEH_NO PER_NO STR_VEH COUNTY DAY MONTH HOUR MINUTE
## <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 1 10001 1 1 1 0 127 1 1 2 40
## 2 1 10002 1 1 1 0 83 1 1 22 13
## 3 1 10003 1 1 1 0 11 1 1 1 25
## 4 1 10003 1 1 2 0 11 1 1 1 25
## 5 1 10004 1 1 1 0 45 4 1 0 57
## 6 1 10005 2 1 1 0 45 7 1 7 9
## # ... with 57 more variables: RUR_URB <int>, FUNC_SYS <int>, HARM_EV <int>,
## # MAN_COLL <int>, SCH_BUS <int>, MAKE <int>, MAK_MOD <int>, BODY_TYP <int>,
## # MOD_YEAR <int>, TOW_VEH <int>, SPEC_USE <int>, EMER_USE <int>,
## # ROLLOVER <int>, IMPACT1 <int>, FIRE_EXP <int>, AGE <int>, SEX <int>,
## # PER_TYP <int>, INJ_SEV <int>, SEAT_POS <int>, REST_USE <int>,
## # REST_MIS <int>, AIR_BAG <int>, EJECTION <int>, EJ_PATH <int>,
## # EXTRICAT <int>, DRINKING <int>, ALC_DET <int>, ALC_STATUS <int>,
## # ATST_TYP <int>, ALC_RES <int>, DRUGS <int>, DRUG_DET <int>, DSTATUS <int>,
## # DRUGTST1 <int>, DRUGTST2 <int>, DRUGTST3 <int>, DRUGRES1 <int>,
## # DRUGRES2 <int>, DRUGRES3 <int>, HOSPITAL <int>, DOA <int>, DEATH_DA <int>,
## # DEATH_MO <int>, DEATH_YR <int>, DEATH_HR <int>, DEATH_MN <int>,
## # DEATH_TM <int>, LAG_HRS <int>, LAG_MINS <int>, P_SF1 <int>, P_SF2 <int>,
## # P_SF3 <int>, WORK_INJ <int>, HISPANIC <int>, RACE <int>, LOCATION <int>
Pipe operator (%>%) from magrittr
## Using multiple object option
per_2015 <-filter(per_2015, INJ_SEV == 4)
p_2015 <- select(per_2015, ST_CASE, PER_TYP, AGE, SEX)
head(p_2015)
## ST_CASE PER_TYP AGE SEX
## 1 10001 1 68 1
## 2 10002 1 49 1
## 3 10003 1 31 1
## 4 10004 1 40 1
## 5 10005 1 24 1
## 6 10006 1 64 1
## Using pipe operator
p_2016 <- per_2016 %>% filter(INJ_SEV==4) %>%
select(ST_CASE, PER_TYP, AGE, SEX)
head(p_2016)
## ST_CASE PER_TYP AGE SEX
## 1 10001 1 37 2
## 2 10002 2 22 1
## 3 10003 1 22 1
## 4 10004 1 36 1
## 5 10005 1 59 1
## 6 10006 1 58 1
acc_veh_2015 <- merge(acc_2015, veh_2015, by = intersect(names(acc_2015), names(veh_2015)), all=TRUE)
acc_veh_2016 <- merge(acc_2016, veh_2016, by = intersect(names(acc_2016), names(veh_2016)), all=TRUE)
## Using multiple object option
acc_veh_2015_clean <- filter(acc_veh_2015, HIT_RUN !=0)
acc_veh_2015_clean <- distinct(acc_veh_2015_clean, ST_CASE, .keep_all = TRUE)
t_2015 <- select(acc_veh_2015_clean, ST_CASE, FATALS)
head(t_2015)
## ST_CASE FATALS
## 1 10008 1
## 2 10035 1
## 3 10055 1
## 4 10069 1
## 5 10128 1
## 6 10146 1
## Using pipe operator
t_2016 <-acc_veh_2016 %>%
filter(HIT_RUN != 0) %>%
distinct(ST_CASE, .keep_all = TRUE) %>%
select(ST_CASE, FATALS)
head(t_2016)
## ST_CASE FATALS
## 1 10146 1
## 2 10203 1
## 3 10275 1
## 4 10398 1
## 5 10401 1
## 6 10408 1
each_dead_2015 <- left_join(t_2015, p_2015, by = "ST_CASE")
each_dead_2016 <- left_join(t_2016, p_2016, by = "ST_CASE")
head(each_dead_2015)
## ST_CASE FATALS PER_TYP AGE SEX
## 1 10008 1 5 38 1
## 2 10035 1 1 91 1
## 3 10055 1 5 63 1
## 4 10069 1 5 51 1
## 5 10128 1 5 51 1
## 6 10146 1 1 30 1
head(each_dead_2016)
## ST_CASE FATALS PER_TYP AGE SEX
## 1 10146 1 5 59 1
## 2 10203 1 2 53 2
## 3 10275 1 5 64 1
## 4 10398 1 5 69 1
## 5 10401 1 5 30 1
## 6 10408 1 5 55 1
## build contigency table of counts of Age
Age_wo_2015 <- table(each_dead_2015$AGE, exclude = NULL)
print(Age_wo_2015)
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
## 2 8 2 3 6 2 1 4 3 3 4 1 6 11 20 23 20 30 29 42
## 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
## 39 48 41 47 37 40 30 37 32 36 23 27 36 25 32 31 34 39 27 30
## 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60
## 29 36 18 24 25 28 28 31 36 42 42 35 35 40 41 35 38 28 26 29
## 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80
## 20 23 14 28 17 18 23 14 12 6 8 15 6 6 9 6 5 4 5 8
## 81 82 83 84 85 86 87 88 89 91 92 93 94 95 998 999
## 3 2 6 7 1 4 5 2 1 1 2 2 2 1 9 9
Age_2015 <- table(cut(each_dead_2015$AGE, breaks = c(0,16,29,59,120)),
exclude = NULL)
Age_2016 <- table(cut(each_dead_2016$AGE, breaks = c(0,16,29,59,120)),
exclude = NULL)
print(Age_2015)
## (0,16] (16,29] (29,59] (59,120] <NA>
## 99 472 957 315 18
print(Age_2016)
## (0,16] (16,29] (29,59] (59,120] <NA>
## 101 556 1070 403 26
Age_year <- as.data.frame(rbind(Age_2015, Age_2016))
colnames(Age_year) <- c("0-16", "16-29", "30-59", "over 60", "NA")
rownames(Age_year) <- c(2015, 2016)
Age_year <- rownames_to_column(Age_year, "Year")
print(Age_year)
## Year 0-16 16-29 30-59 over 60 NA
## 1 2015 99 472 957 315 18
## 2 2016 101 556 1070 403 26
2. ggplot2 for data visualization
library(wesanderson) # load color palette
Age_year[, "NA"] <- NULL
# reshpae to long format
Age_reshape <- gather(Age_year, Age, Counts, 2:5)
print(Age_reshape)
## Year Age Counts
## 1 2015 0-16 99
## 2 2016 0-16 101
## 3 2015 16-29 472
## 4 2016 16-29 556
## 5 2015 30-59 957
## 6 2016 30-59 1070
## 7 2015 over 60 315
## 8 2016 over 60 403
p <- ggplot(Age_reshape)+
geom_bar(mapping = aes(x = Age, y = Counts, fill = Year), width = 0.5,
position = "dodge", stat="identity")+
coord_cartesian(ylim=c(0,1200))+
theme_light()+
xlab("Age Range")+
ylab("Fatality Counts")+
ggtitle("Fatalities by Age")+
theme_light()+
theme(legend.position="bottom", legend.direction="horizontal",
legend.title = element_blank())+
scale_fill_manual(values=wes_palette(name="Chevalier1"))+
theme(axis.text.x=element_text(angle=45,hjust=0.5, vjust=0.4),
plot.title = element_text(hjust = 0.5))
print(p)